I knew R was versatile, but DANG, people do a lot with it:
> > … I don’t think anyone actually believes that R is designed to make *everyone* happy. For me, R does about 99% of the things I need to do, but sadly, when I need to order a pizza, I still have to pick up the telephone. —Roger Peng
> There are several chains of pizzerias in the U.S. that provide for Internet-based ordering (e.g. www.papajohnsonline.com) so, with the Internet modules in R, it’s only a matter of time before you will have a pizza-ordering function available. —Doug Bates
Indeed, the GraphApp toolkit … provides one (for use in Sydney, Australia, we presume as that is where the GraphApp author hails from). —Brian Ripley
So, heads up: the following post is super long, given how much R was covered at the conference. Much of this is a “notes-to-self” braindump of topics I’d like to follow up with further. I’m writing up the invited talks, the presentation and poster sessions, and a few other notes. The conference program has links to all the abstracts, and the main website should collect most of the slides eventually.
Wednesday was the first day of the conference proper. I enjoyed my morning walk from the hotel through a grove of lush trees; as Frank Harrell (Biostats department chair, and author of Hmisc) pointed out in his welcome speech, the campus is a nationally designated arboretum.

Di Cook started us off with the warning that Every Plot Must Tell A Story – Even In R. Many R packages have few or no examples for the data, and even when they do, plots of the data are scarce or often unhelpful. She laid out a wishlist: packages should come with example plots that display the raw data (showing blatant outliers or clusters), then explore its structure, and then illustrate how the package’s tools can be used for modeling. Given a new dataset, her default first checks include side-by-side boxplots, parallel coordinate plots, or just throwing it all into GGobi. I also liked her advice that it’s easier to make comparisons to a vertical or horizontal line than to a diagonal one. I can’t find her slides but here’s a mockup:
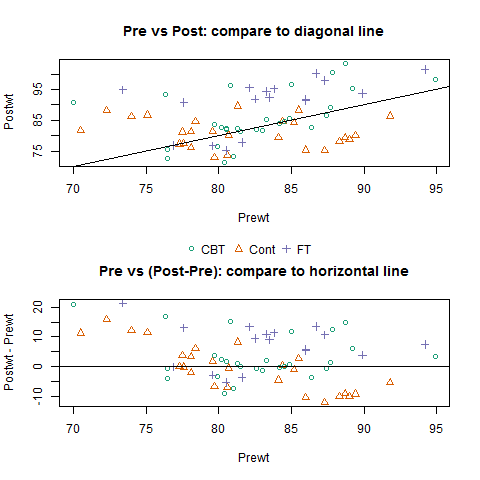
However, I was surprised at a few of her color choices! When discussing species of crabs with colors in the name (Blue Crabs and Orange Crabs), I would have used those colors in the scatterplots too, to help readers remember which is which. And when separating them out by both species and sex, I’d have kept colors consistent with previous plots. Instead, one plot had pink females and blue males; I believe the next had pink for Female Blue Crabs, but blue for Female Orange Crabs, etc… There’s a lot of scope for confusion there!
Audience members also pointed out that it’s good to choose colors that are colorblind-safe. ColorBrewer has good starting suggestions; you can also test out your graphics by running them through Vischeck or ColorOracle.
On Wednesday afternoon, Bryan Hanson gave an invited talk on HiveR. I’d heard about hive plots before, but Hanson’s talk really clarified for me exactly what they are and how they differ from more traditional network diagrams. Very loosely speaking: each node is a thing; things of different types are on different axes; things that are related are connected by lines. For example, pollinating insects and plants are two types of things, so they would be nodes on two different axes, and connecting lines show which insects pollinate which plants. That’s simple enough, but add more dimensions (more categories of things, i.e. more axes) and hive plots start to display patterns in the data much more clearly than network diagrams can. Also, since the algorithms that space out network diagrams can get quite messy, removing a node from a network diagram may cause it to be redrawn in a totally different way… but removing a node from a hive plot leaves everything else as it was, making across-plot comparisons possible. Thinking of hive plots as radial parallel-coordinate plots gave me some ideas for how they might be useful in my own work.
Hanson was followed by Simon Urbanek on web-based interactive graphics and R in the cloud. Classically, an analyst’s data and R have been local to their machine, but we need to start thinking about workflows that allow for distributed data, sharing analyses, reproducibility, and remote computing. He demo’ed what looked like an interactive R notebook; I couldn’t tell if it’s a package in the works or just a tool for his own use. I hope his slides are posted soon, since the amount of material was overwhelming and I’d like to look it over more slowly. He mentioned many useful packages and other tools including iplots, iPlots eXtreme, Rserve, RSclient, FastRWeb, RCassandra, WebGL, facet…
Thursday’s invited speakers included Norm Matloff, on parallel computation in R. His talk went outside my area of expertise, but I learned that most parallel computing works on “embarrassingly parallel” (EP) problems, i.e. ones that are very simple to break into chunks and then combine at the end. He gave some pointers for dealing with non-EP problems and presented a few new tools for this, summarized in his slides. I’d also like to read his online book on this topic.
The next speakers were RStudio‘s JJ Allaire and Iowa’s Yihui Xie, on reproducible research in R. Not only should our code be reproducible, but our documents too — we don’t want to cut-and-paste the wrong table or graph into our papers. Many of us are familiar with Sweave, a tool for weaving R code and results directly into a LaTeX document… but JJ and Yihui showed how RStudio and the knitr package make this even easier than in Sweave. As JJ put it, we want to “take the friction out of the process of building these documents.”
For example, Yihui pointed out that if you copy-and-paste from usual R output into a new script, you have to delete or comment out the results, and remove the “>” prompt symbol from commands, before you can re-run the code… But knitr defaults to printing the commands with no prompt and the results commented out, so you can paste them directly back into R. Brilliant!
Also, as Yihui advised the academics in the audience, not all of your students will go into research but they all have to do homework — so train them well by requiring reproducible homework assignments, and those who do research later will be well-prepared.
This talk also introduced me to Markdown and MathJax (alternatives to or tools for HTML and ?), and to the RPubs site for “easy web publishing from R.”
Thursday afternoon ended with a discussion (see the slides and code) of other languages R users should know about. This included a “shootout” comparing the ease of writing and speed of running a Gibbs sampler in several languages, inspired by a similar comparison on Darren Wilkinson’s blog.
- Dirk Eddelbuettel voted for C++, saying that we need “barbells not bullets” — no single language will do everything, so we should pick a few that have complementary strengths. Dirk said that
RcppandRInsidemake it easier to cross between R and C++ than it is to call C using base R, while still giving similar speed benefits. - Doug Bates gave an introduction to the new (6-month-old?) language Julia, which seems to have R- or MATLAB-like syntax but can run quite a bit faster, either interactively or compiled. Julia seems to lack some of R’s nice features like native handling of NAs, default argument values for functions, named arguments, and dataframes (though Harlan Harris, of DC’s R Users Meetup group, got a shoutout for his work on the latter). Julia shows a lot of promise but its developers clearly have a lot of work left to do… including convincing Wikipedia of its importance!
- Chris Fonnesbeck’s Python demo was quite convincing. Python beats R at iterating through for-loops, which is necessary for things like Gibbs sampling; it’s a “glue language” that connects well to other languages like C or C++; and it’s high-level and readable: people call it “runnable pseudocode” and apparently its syntax is easy to remember. He ran his demo in IPython, which seemed to be an interactive notebook that runs in a web browser and allows for interactive graphics, parallel computing, etc… but can also be saved as a static document and emailed to someone. I really want to try this out and compare it to RPubs.
Julia did seem to beat the simplest Python or Rcpp code in the Gibbs-sampler shootout, but all three languages allow for tweaks that made the final versions pretty comparable. (The term “shootout” is cute — I might steal it for describing model comparisons in my future talks.)
Also, if I understood correctly, R and the rest of these languages all rely on the same linear algebra libraries. It’s possible to replace the libraries that shipped with your computer and install faster versions that’ll speed up each of these languages. I scribbled down “BLAS/LINPACK/openBLAS?” in my notebook and clearly need to follow up on this 🙂
Friday morning, Tim Hesterberg of Google presented his dataframe and aggregate packages. Tim found some inefficiencies in R’s base code for dataframes (i.e. they copy the dataframe several times when replacing elements), so dataframe basically replaces them with new code: the user can still call dataframes the same way, but now they run up to 20% faster. R-core members advised caution, since the low-level functions used in his speedups can be dangerous … but Hesterberg hasn’t seen any problematic results so far. Also, his aggregate package speeds up some common aggregation and tabulation tasks: taking means, sums, and variances of rows or columns, perhaps by factors, allowing for weights, etc. I was amused to hear he was inspired by SAS’s PROC SUMMARY and its use of BY statements; he may have been the only presenter to mention SAS in a positive light…
Finally, Bill Venables closed the conference with an invited address on “Whither R?” (slides, paper). He admitted that he converted Frank Harrell and Brian Ripley to R, “so I’m to blame” 🙂 In his overview of R’s history, he said that with earlier tools, “you were spending most of your ingenuity getting the tool to *do* the computation.”
Bill compared John Tukey’s view, that statistics work is detective work, to how many schools teach it as “judge and jury work: stand on the side saying ‘ah-ah, 5%, go back and do it again…'” Also, in his experience, the difference between applied math and statistics is the attitude towards the data: applied mathematicians build a clear model, then desperately look for data to calibrate it, while statisticians collect any data they can to get an insight into the process and respect whatever is going on.
He presented an example of statistical analyses for the Australian shrimp fishery, with some funny quotes. On the difference between land and ocean ecologists: “Counting fish is a bit like counting trees, except that they’re under water and they move.” And on the difference between Tiger prawns and Brown prawns: “It’s easy to remember which is which, because the Brown one is less brown.” I liked his choice of spatial coordinates (instead of lat and long, one coordinate was distance on a line along the coastline and the other was distance from shore). It was also instructive to see a success story where a model gave useful predictions *but also* justified a new project with data collection to check on the model’s stability over time.
Bill also classified R packages into four types: wrong or inept packages; packages for “refugees,” easing the transition from another software; “empires” or interlocking suites of packages with a different philosophy than base R; and GUIs, which “ease the learning curve but lead to a dead end only part of the way up the hill.”
Finally, although Bill brought up the pizza quotes atop this post, he said that R doesn’t do everything (and may be nearing its boundaries, programming-wise) so it’s worth learning other complementary tools. However, it’s a great training tool to get people hooked on interactive data analysis and the detective work of stats, and it has a bright future for data analysis and graphics, assuming an explicit succession plan can be worked out for the R-core team.
Next, some highlights from the presentation and poster sessions:
- Ben Nutter on
lazyWeave. Telling SAS users to switch to R and Sweave means learningtoo, i.e. two languages at once, which is daunting.
lazyWeavedeals with the, so they can get by with just knowing R. He promised
lazyHTMLandlazyBeamerpackages soon as well. - Eric Stone on
audiolyzR, the first plausible example I’ve seen of “data sonification” (the auditory equivalent of data visualization). With his demo of “training plots” it was easy to see how each sonification corresponded to high positive correlation, negative correlation, outliers, and random noise. - Heather Turner on using
gnmand Diagonal Reference Models to analyze social mobility in the UK. These models seem to be a useful way to summarize trajectories (such as changes in social class) while avoiding some common oversimplifications.
(Heather was also the chair for my session on official statistics. She uses data from a large survey linked to administrative records, much as I do in my work; but since she’s not in a national statistical office, she can’t access the data directly — she has to email them her code and wait for results to be sent back. What a workflow!) - Mine Cetinkaya-Rundel on using R for introductory statistics. She’s contributing to an open-source statistics textbook, using R and RStudio to teach statistics and a little programming too. They’ve found that writing the code helps students to understand simulations, sampling distributions, confidence, bootstraps, etc. much better. However, for classical inference, they also give the students a unified function that connects topics that may otherwise seem disjoint. Their functions also provide a lot more output by default than R usually does, more like SAS or SPSS; and they try to throw “human-readable error messages” 🙂
- Yihui Xie on
cranvas, a powerful and fast graphics package that easily allows interactions such as linking and brushing. Marie Vendettuoli also usedcranvasto show us some hammock plots, i.e. parallel coordinate plots with line thickness as another variable. Unfortunately it only works on Linux so far, and the graphics only run inside of R (i.e. you cannot save or export them as interactive objects yet). - Jae Brodsky on using R at the FDA with regulated clinical trials. The R Foundation has put out a document on guidelines for using R in a regulatory compliance setting.
- Wei Wang on
mrpfor Multilevel Regression and Poststratification a.k.a. “Mr P.” I need to look into this some more: it looks very similar to the small area estimation work I do, wrapped up in slightly different language. - Edwin de Jonge on
tabplotfor tableplots, a nifty way to summarize very large unit-level datasets such as a large survey or a census. He’s also working on a new version that uses the D3 JavaScript library to make the tableplots interactive; I’d love to see how that happens, so perhaps I can use R to help populate my D3 maps. - Anthony Damico on
SAScii, for parsing SAS input files and auto-translating them into R code. Some public datasets are published as text files with SAS input instructions, such as the Census Bureau’s Survey of Income and Program Participation (SIPP). SAScii helps read such datasets into R, although it can take many hours for large datasets like SIPP. - Bob Muenchen on helping your organization migrate to R. As the author of resources like R for SAS and SPSS Users, he had many good tips on how to sell R without alienating people used to other software. Show them how little R they need to get by at first; transition by phases; try some GUIs like DeduceR and SAS-to-R conversion resources like Rconvert.com. Bob’s website hosts his slides and other good materials.
- In my own session (on official statistics) the other talks were by Jan-Philipp Kolb, on synthetic universes for microsimulation; and Mark van der Loo, on editing and correcting survey data.
Kolb’s group work with indicators of poverty and social exclusion for the AMELI project. They createdsimPopulationandsimFramefor generating synthetic datasets that mimic the properties of their real data, so that other analysts can do research and reproduce their results without having to be granted access to the real data (which contains private information and cannot be shared). They also organized their data into a “synthetic map” which is a new and interesting concept to me.
Van der Loo presented tools for automated editing, correction, and imputation of mistakes or gaps in survey data. His toolseditrulesanddeducorrectmake it easy to input the rules (i.e. ages must be nonnegative, unemployed have a salary of 0, etc.), and they will flag or correct the mistakes or missing data that can be derived from the remaining data. I like his term “error localization” for finding the fewest number of fields that would have to be adapted so that no rules are violated. - Przemysław Biecek on parallel R with
nzrandnpsRcluster, and Antonio Piccolboni onRHadoopandrmr. Both discussed frameworks for processing big data on clusters; Biecek had a great example of sentiment analysis of tweets on soccer matches. - Andreas Alfons on
cvToolsfor cross-validation and model selection. This package makes it easier to cross-validate and compare fitted models on your choice of loss function. If it already has a methor for your model type, such as regression (lm) or robust regression (lmrob), it’s easy to give it the fitted model, desired number of cross-validations, and prediction loss function to use. If not, developers can write wrappers for new model types. He’s also working on extensions for the next iteration, called Perry (for Prediction ERror in Regression models?) - Karl Millar on scaling up R to the kind of multi-terabyte datasets used at Google. Their data is so high-dimensional that you’d lose information if you just analyzed a subsample; and even small coefficients will have real importance on events that happen over and over. He gave an overview of MapReduce, their approach to embarrassingly-parallel problems, and of Flume, a wrapper/abstraction on top of MapReduce that’s only about 10% slower but much easier to work in. When an audience member asked how we can get to play with these tools, Millar responded, “Send us job applications” 🙂
It seems that Google has about 200 regular R users and another 300 part-timers. They use R a lot for internal analyses, but not for real-time production code.
While in Nashville I also got to meet Nicholas Nagle, from the University of Tennessee in Knoxville, whose interactive map I’ve linked to before. He pointed me to some new resources for spatial statistics including MapShaper, an online tool for simplifying boundaries on shapefiles.
Finally, I was pleased to learn that R can be installed onto a USB stick and run from there directly, without installing anything on the host computer. This is very useful when, say, your presentation requires a live R demo but you realize at the last minute that your brand-new laptop has no VGA socket for connecting to the projector… not that I would know… *cough* Seriously, this was a life saver. Portable Firefox came in handy too, for demo’ing SVG-based graphics that didn’t work in the presentation computer’s old version of Internet Explorer.
And although all the talks I attended were solid, I found some great advice for the future, by Di Cook and Rob Hyndman, on giving useR talks.
A huge thank-you to the organizers, Vanderbilt, Nashville, and all 482 attendees. I hope to make it to useR 2013 in Albacete, Spain!

Related links:
- My post on the Tuesday tutorials, and the slides and code from my own talk
- Conference highlights from others: Stephanie Kovalchik, David Smith, Gregory Park, Steve Miller, and Pairach Piboonrungroj


Very good review! It makes me really sorry that I didn’t make it this year. Thanks for putting a link back to my post too.
I’m glad that you like the idea of removing > by default. Some useRs are so used to it that it is difficult to convince them this is a horrible thing for readers. Journal of Statistical Software recommends R> as the prompt, and this is the single reason that I hate it because each time I read a JSS paper, I have to try hard to strip off the stupid prompts before I run the code (I love anything else about JSS).
Pairach: Thanks for all your links too, and sorry you couldn’t make it — but next year, I hope!
Yihui: Absolutely. Statistics is complicated enough already — I’m a huge fan of anything that removes needless friction from my work. Thanks for all your great demos and talks that week!